| Journal of Endocrinology and Metabolism, ISSN 1923-2861 print, 1923-287X online, Open Access |
| Article copyright, the authors; Journal compilation copyright, J Endocrinol Metab and Elmer Press Inc |
| Journal website https://jem.elmerpub.com |
Short Communication
Volume 15, Number 5, December 2025, pages 220-226
Altered CTLA-4 Expression and Genetic Variation Drive Autoimmune Susceptibility in Turner Syndrome
Aldianne Barbosaa, Raysa Laranjeiraa, Thays Lucenaa, b, Adriana Bispoc, Luana Santosa, Juliana Arcoverdea, Carla Santosa, Camilla Limab, Andrea Duarted, Barbara Gomese, Natassia Bufalof, Jaqueline de Azevedo-Silvaa, b, Laura Wardf, Neide Santosa, g
aDepartamento de Genetica, Universidade Federal de Pernambuco, Recife, PE, Brazil
bLaboratorio de Imunopatologia Keizo Asami - iLIKA, Universidade Federal de Pernambuco, Recife, PE, Brazil
cInstituto Federal do Sertao Pernambucano - Campus Serra Talhada, PE, Brazil
dServico de Genetica Medica, Instituto de Medicina Integral Professor Fernando Figueira, Recife, PE, Brazil
eServico de Endocrinologia Pediatrica do Hospital das Clinicas, Universidade Federal de Pernambuco, Recife, PE, Brazil
fLaboratory of Cancer Molecular Genetics, School of Medical Sciences (FCM), University of Campinas (UNICAMP), Campinas, Sao Paulo, Brazil
gCorresponding Author: Neide Santos, Departamento de Genetica, Universidade Federal de Pernambuco, 50740-600 Recife, Pernambuco, Brazil
Manuscript submitted October 6, 2025, accepted November 10, 2025, published online December 24, 2025
Short title: CTLA-4 Expression and Autoimmunity in TS
doi: https://doi.org/10.14740/jem1578
| Abstract | ▴Top |
Background: Patients with Turner syndrome (TS) exhibit an increased susceptibility to autoimmune and chronic inflammatory diseases compared to the general female population. Recent studies have implicated immune regulatory genes, including cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), a key player in immunosuppression, in this predisposition. This study investigated the CTLA-4 rs3087243 (G>A) polymorphism and gene expression profiles in patients with TS to assess their potential association with immunological dysregulation.
Methods: A genetic association study was conducted in 112 patients with TS (with and without autoimmune/inflammatory diseases) and compared to 241 healthy women using TaqMan genotyping. Additionally, CTLA-4 mRNA levels were quantified via quantitative polymerase chain reaction (qPCR) in 15 TS patients and 15 controls, including TS subgroups with (n = 9) and without (n = 10) autoimmune/inflammatory conditions.
Results: The G allele was more prevalent in TS patients with autoimmune diseases (P = 0.047; odds ratio (OR) = 0.51; 95% confidence interval (CI) = 0.25 - 1.02), while the GA genotype appeared protective in controls (P < 0.001; OR = 0.14; 95% CI = 0.06 - 0.3). No association was found between CT60 and inflammatory diseases in TS. Notably, CTLA-4 expression was significantly downregulated in patients with TS compared to controls (-19.38-fold change; P < 0.001), although no differences were detected between TS subgroups with and without immune-related conditions (P = 0.8413).
Conclusions: These findings highlight a possible role of CTLA-4 in TS-associated immune dysregulation, particularly through reduced expression, which may contribute to altered immune responses. Although the rs3087243 polymorphism showed limited clinical association, its differential distribution in patients with TS versus healthy controls underscores the genetic complexity of immune regulation in TS. Further research is needed to elucidate the mechanistic links between CTLA-4 variants, expression patterns, and autoimmune susceptibility in TS.
Keywords: Autoimmune diseases; Inflammatory diseases; CTLA-4; Gene expression; rs3087243; Immune dysregulation; Turner syndrome
| Introduction | ▴Top |
Turner syndrome (TS) is a chromosomal disorder caused by the partial or complete loss of one X chromosome, occurring in approximately 1 in 2,500 live female births [1, 2]. It is the only monosomy compatible with life and typically results from chromosomal non-disjunction. The most common karyotype, 45,X, accounts for 40-60% of cases, whereas mosaicism (e.g., 45,X/46,XX) and structural X chromosome abnormalities (e.g., isochromosomes and deletions) occur in 30% of patients. Additionally, 10-12% of individuals harbor a second lineage with Y chromosome material [3, 4].
Common phenotypic features include short stature, gonadal dysgenesis, pubertal delay, ovarian insufficiency, and infertility, along with characteristic physical findings, such as a short/webbed neck, wide chest, and cubitus valgus. These patients also face an elevated risk of endocrine, cardiovascular, and autoimmune disorders, including Hashimoto’s thyroiditis, type 1 diabetes, celiac disease, and rheumatoid arthritis [3-5].
Patients with TS have approximately twice the risk of developing autoimmune disease (AID) and chronic inflammatory disease (CID) compared to the general female population, establishing these conditions as hallmark clinical features of the syndrome [6]. Hashimoto’s thyroiditis is the most prevalent autoimmune manifestation, while other frequently associated conditions include type 1 diabetes mellitus, celiac disease, inflammatory bowel diseases (including Crohn’s disease and ulcerative colitis), juvenile rheumatoid arthritis, and endocrine disorders such as Addison’s disease [7, 8]. Additional autoimmune comorbidities include autoimmune hepatitis, psoriasis, vitiligo, and alopecia. Despite well-documented clinical associations, the underlying immunological mechanisms remain poorly understood, with particularly limited investigation of immune-related genetic factors in patients with TS [9, 10].
Immune regulatory genes, particularly cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), have been implicated in various diseases [8]. Located on chromosome 2q33, CTLA-4 encodes a critical cell surface protein that functions as a competitive inhibitor of CD28 receptor binding to CD80/CD86 ligands on antigen-presenting cells, thereby suppressing T-cell activation and proliferation [11, 12]. Predominantly expressed in activated CD4+ and CD8+ T cells, this immune checkpoint molecule serves as a fundamental negative regulator of immune response. Through its inhibitory function, CTLA-4 maintains peripheral tolerance, prevents autoimmunity, and contributes to immune system homeostasis [11, 13, 14].
This study investigated the CTLA-4 rs3087243 (CT60) polymorphism in patients with TS, evaluating its association with AID and CID, and analyzed CTLA-4 gene expression to explore its potential contribution to immune dysregulation in TS. Through a comprehensive evaluation of these genetic variations and their immunoregulatory consequences, we sought to elucidate the molecular mechanisms underlying the heightened susceptibility to autoimmune and inflammatory disorders characteristic of TS.
| Materials and Methods | ▴Top |
Study design and participants
This cross-sectional study aimed to provide a comprehensive cytogenetic and molecular characterization of patients diagnosed with TS between 2006 and 2018. The participants were recruited from the Pediatric Endocrinology Service at Hospital de Clinicas, Universidade Federal de Pernambuco (UFPE) and the Medical Genetics Service at Instituto de Medicina Integral Professor Fernando Figueira (IMIP). Clinical data were extracted from medical records.
Inclusion criteria required a cytogenetic diagnosis of TS. Individuals with 46,XX or 46,XY karyotypes were excluded. The healthy control (HC) group consisted of individuals of the same sex, with normal karyotypes and without autoimmune, endocrine, or inflammatory disorders. Controls for gene expression analyses were recruited from the community and completed a questionnaire to confirm eligibility.
Ethical considerations
This study received ethical approval from the Research Ethics Committee of Health Sciences Center at Universidade Federal de Pernambuco (UFPE) (approval number: CAAE 0485.0.172.000-11) and IMIP (approval number: 802/06). Written informed consent was obtained from all participants or their legal representatives, following the guidelines of the Declaration of Helsinki.
DNA extraction, selection of SNPs, and genotyping
Genomic DNA was extracted from peripheral whole blood samples using the Illustra™ Blood GenomicPrep Mini Spin Kit (GE Healthcare, Little Chalfont, UK) following the manufacturer’s standardized protocols. The extracted DNA samples were subsequently stored at -20 °C until further analysis to maintain genomic integrity.
For genotyping analysis, the CTLA-4 gene single nucleotide polymorphism rs3087243 (G>A transition, assay ID: C_3296043_10) was specifically selected based on its minor allele frequency (MAF < 10%) in the target population. The genotyping procedure was performed using commercially available TaqMan® SNP Genotyping Assays (Applied Biosystems, Life Technologies, Carlsbad, CA, USA) and analyzed on an ABI PRISM® 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s recommended thermal cycling conditions.
The CT60 polymorphism (rs3087243) was analyzed using a systematic two-stage approach. In the first stage, 24 TS patients diagnosed with AIDs were compared with 88 TS controls without autoimmune manifestations. The second stage involved a comparative analysis between 22 patients with TS presenting with CIDs and 90 TS controls lacking such pathological conditions. To provide a broader context, an additional comparative analysis was performed between the combined TS patient cohort (n = 112) and an HC group (n = 241) previously characterized by Bufalo et al [15]. The HC group was deliberately selected to comprise exclusively female participants, with a mean age of 36.86 years (standard deviation (SD) ± 12.95), ensuring appropriate sex matching with the TS study population.
RNA extraction, cDNA synthesis, and gene expression assay
Total RNA was extracted from peripheral blood leukocytes using TRIzol® Reagent (Invitrogen, USA) according to the manufacturer’s protocol. Prior to cDNA synthesis, the RNA concentration was standardized to 500 ng per sample to ensure consistency across all the experimental conditions. Complementary DNA (cDNA) synthesis was subsequently performed using the ImProm-II™ Reverse Transcription System (Promega, USA) following the manufacturer’s specifications.
Gene expression analysis was conducted using TaqMan® probes specifically designed for the target gene CTLA-4 (Hs.00175480, Thermo Fisher Scientific). Relative gene expression levels were normalized using two endogenous reference genes: hypoxanthine phosphoribosyltransferase 1 (HPRT1) and beta-2-microglobulin (B2M) (Thermo Fisher Scientific). These reference genes were analyzed using the SYBR Green detection system with their respective primer sequences: HPRT1 (forward: 5'-ACAGGACTGAACGTCTTGCT-3', reverse: 5'-GAGCACACAGAGGGCTACAA-3') and B2M (forward: 5'-TGAGTGGCATGAAGAAGGTGT-3', reverse: 5'-GGCAGTTCTTTGCCCTCTCT-3') (Thermo Fisher Scientific). The selection of these endogenous reference genes was based on their demonstrated expression stability across the tested samples and experimental conditions, as validated by the GeNorm analysis. All quantitative PCR assays were performed using an ABI 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Gene expression analysis was conducted based on quantification cycle (Cq) values, and relative mRNA levels were calculated using the 2-ΔΔCt normalization method [16], following the quality criteria established by MIQE guidelines [17].
CTLA-4 gene expression analysis encompassed two distinct comparative frameworks. The primary analysis involved 15 patients with TS serving as the case group (mean age 21.07 ± 9.33 years) compared with 15 healthy female volunteers constituting the control group (mean age 30 ± 10.01 years). Subsequently, a secondary analysis was conducted to examine intragroup differences within the TS population, comparing nine TS patients diagnosed with AIDs and/or CIDs (mean age 21.67 ± 8.26 years) with 10 TS patients without such clinical manifestations (mean age 21.3 ± 7.89 years), thereby serving as an internal control group.
Statistical analysis
Statistical analysis for the association study was conducted using a comprehensive approach employing multiple analytical platforms. Genotype and allele frequency analyses were performed using the SNPStats web-based tool [18] in conjunction with R statistical software version 4.1.2 [19].
Hardy-Weinberg equilibrium (HWE) assessment and determination of allele and genotype frequencies were evaluated using the Chi-square test (χ2). Statistical significance for differences in allelic and genotypic distributions between groups was determined using Fisher’s exact test, with statistical significance set at P < 0.05. To ensure adequate statistical power, post-hoc power analysis was conducted using G*Power software (version 3.1.9.7 [20]), with a type I error probability (α) of 0.05.
For gene expression analyses, data normality was assessed using the Shapiro-Wilk test to determine the appropriate statistical approach. Subsequently, either Student’s t-test (for parametric data) or the Mann-Whitney U test (for non-parametric data) was applied, maintaining a significance threshold of P < 0.05. All statistical computations were performed using R statistical software, and data visualization and graphical representations were generated using GraphPad Prism software version 8.0 (GraphPad Software, San Diego, CA, USA).
| Results | ▴Top |
Clinical features
The study cohort comprised 112 TS patients (mean age at diagnosis: 12 ± 8.47 years). Table 1 presents a comprehensive overview of the clinical characteristics of patients with TS. The analysis revealed that 21.43% of patients developed at least one AID, including Hashimoto’s thyroiditis, hypothyroidism, and alopecia areata. Furthermore, 19.64% of patients had CIDs, including obesity, chronic otitis media, dermatitis, persistent allergic rhinitis, and recurrent tonsillitis.
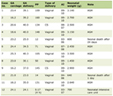 Click to view | Table 1. Clinical Features of Patients With Turner Syndrome |
Notably, the clinical presentation was complex, as several patients exhibited concurrent manifestations of both autoimmune and inflammatory conditions. The overall prevalence of immunological disorders was substantial, affecting approximately one-third of the study population (34% of 112 patients).
The distribution of immunological complications was as follows: 14.29% (n = 16) presented exclusively with AIDs, 12.5% (n = 14) exhibited solely inflammatory conditions, and 7.14% (n = 8) demonstrated concurrent autoimmune and inflammatory manifestations. These findings underscore the significant burden of immune-related complications in patients with TS and highlight the multifaceted nature of immunological dysfunction associated with this chromosomal disorder.
CTLA-4 polymorphism in TS patients with AIDs
The genotype and allele frequencies for the CTLA-4 rs3087243 (G>A) polymorphism were systematically compared between patients with TS presenting with AIDs as the case group and patients with TS without AIDs serving as the control group (Table 2). Statistical analysis revealed a significantly higher frequency of the G allele among patients with TS and AIDs (P = 0.047; OR = 0.51; CI = 0.25 - 1.02), indicating a distinctive allelic distribution pattern in this patient subgroup. Conversely, the comparison of genotypic distributions between the two groups did not yield any statistically significant differences. The study maintained a robust statistical power of 97% with a 5% confidence level. Notably, the observed genotype frequencies deviated from the HWE expectations for the evaluated single nucleotide polymorphisms (SNPs).
 Click to view | Table 2. Genotype and Allele Distribution of CTLA-4 Gene Polymorphism in TS Patients With AIDs (Case Group), TS Patients With CIDs (Case Group), and TS Patients (Control Group) |
CTLA-4 polymorphism in TS patients with CIDs
Table 2 presents a comprehensive analysis of the CTLA-4 rs3087243 (G>A) polymorphism distribution among TS patients with CIDs serving as the case group, compared to TS patients without CIDs serving as the control group. Comparative analysis revealed no statistically significant differences in either allelic or genotypic frequency distributions between the two groups. The study maintained a robust statistical power of 93% with a predetermined significance threshold of α = 0.05. Consistent with the previous AID analysis, the observed allelic and genotypic distributions of the evaluated SNPs significantly deviated from HWE expectations. The deviation from HWE in the TS cohort may reflect the genetic admixture characteristic of the Brazilian population.
CTLA-4 polymorphism in TS patients compared to the HC group
Table 3 presents a comparative analysis of the CTLA-4 rs3087243 (G>A) polymorphism between patients and HCs. While no statistically significant differences were detected in allelic distribution between the two groups, a notable and significant difference was observed in genotypic distribution (P < 0.001; OR = 0.14; CI = 0.06 - 0.3). These findings suggest that the GA genotype may confer a protective effect in the HC population compared with TS patients. The analysis demonstrated optimal statistical power at 100% with a 5% confidence level, ensuring the robust detection of true associations.
 Click to view | Table 3. Genotype and Allele Distribution of CTLA-4 Gene Polymorphism in TS Patients and Healthy Control Group [15] |
A critical observation from this analysis is the differential adherence to the HWE between the study populations. Notably, the SNP distributions in patients with TS deviated significantly from the HWE expectations, whereas the HC group maintained equilibrium. This deviation in the TS cohort may reflect underlying population stratification, selection pressures, or the complex genetic architecture inherent to TS and warrants careful consideration in the interpretation of these genetic association results.
CTLA-4 gene expression assay
CTLA-4 mRNA levels were assessed in patients with TS and controls using relative gene expression analysis. The initial comparative analysis revealed that patients with TS exhibited significantly diminished CTLA-4 expression, demonstrating a substantial 19.38-fold downregulation compared to HCs (P < 0.001, P = 3.073 × 10-14) (Fig. 1a). Conversely, when examining CTLA-4 expression levels between patients with TS presenting with AIDs and/or CIDs (n = 9) and those without such comorbidities (n = 10), no statistically significant differences in gene expression were detected (fold change = 1.0207, P = 0.8413) (Fig. 1b).
 Click for large image | Figure 1. Differential CTLA-4 gene expression in TS: (a) patients TS vs. HCs (-19.38 FC; P < 0.001); (b) comparative analysis of patients with AIDs and/or CIDs (case group) versus TS without such conditions (control group) (-1.0207 FC; P = 0.8413). AID: autoimmune disease; CID: chronic inflammatory disease; FC: fold change; HC: healthy control; TS: Turner syndrome. |
| Discussion | ▴Top |
In the present study, 34% of the sample had some form of autoimmune and/or inflammatory disease, consistent with findings from previous studies [6, 21]. However, it is important to note that the association between a specific karyotype and the development of immunological conditions in TS remains controversial [8].
Our study provides novel insights into the role of CTLA-4 in immune dysregulation among patients with TS, focusing on the rs3087243 polymorphism and gene expression patterns. These findings underscore the genetic complexity of immune regulation in TS and its potential contribution to autoimmune and inflammatory susceptibility.
The higher prevalence of the G allele in patients with TS and AIDs (P = 0.047) suggests a possible association with immune dysfunction, aligning with previous studies linking CTLA-4 polymorphisms to autoimmune conditions such as Graves’ disease and rheumatoid arthritis [22, 23]. Notably, the GA genotype exhibited a protective effect in HCs (P < 0.001), a phenomenon absent in patients with TS, likely due to the unique genetic and hormonal milieu of TS. This discrepancy highlights the distinct immunogenetic landscape of the syndrome, which may disrupt typical regulatory mechanisms [5].
The significant downregulation of CTLA-4 expression in patients with TS (-19.38-fold, P < 0.001) further supports its role in immune dysregulation. CTLA-4 is a critical checkpoint molecule for maintaining peripheral tolerance, and its reduced expression can impair immune homeostasis, predisposing patients with TS to autoimmunity [11, 12]. However, the lack of differential expression between TS subgroups with and without AIDs or CIDs suggests that CTLA-4 downregulation is a generalized feature of TS cells rather than a specific marker for these conditions. This finding contrasts with studies on other autoimmune diseases, such as systemic lupus erythematosus, where CTLA-4 dysregulation manifests differently [24], emphasizing the need for syndrome-specific investigations.
The absence of HWE in the TS cohort for rs3087243 may reflect the population stratification typical of the highly admixed Brazilian population or selection pressures inherent to TS [25]. While this deviation complicates genetic association analyses, it underscores the importance of considering TS-specific genetic architectures in future studies. Our results, combined with prior work on CTLA-4 polymorphisms in TS [9], suggest that distinct variants may contribute differentially to immune dysregulation depending on their genomic context and functional impact.
This study presents some limitations, including limited screening for certain AIDs, such as celiac disease (CD), the small sample size for gene expression analyses, and the deviation from HWE. It is important to note that, in our cohort, only a few patients with TS underwent screening for CD, and considering its potential for subtle and nonspecific presentation, underdiagnosed is likely, as described in another study [26]. Nevertheless, these results provide a basis for future research exploring additional immunological regulators, longitudinal outcomes, and potential therapeutic targets in TS.
In conclusion, our study highlights the multifaceted role of CTLA-4 in TS-associated immune dysregulation driven by both genetic variants and expression alterations. However, the interplay between CTLA-4 and other immune regulators remains unclear. Further research should explore the mechanistic pathways and potential therapeutic targets to mitigate immune-related complications in TS.
Acknowledgments
The authors thank the patients, parents, and physicians for their cooperation in obtaining the data used in this research.
Financial Disclosure
The present research was supported by Fundacao de Amparo a Ciencia e Tecnologia do Estado de Pernambuco (FACEPE) (grant number APQ-0638-2.02/12), Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq), Empresa Brasileira de Servicos Hospitalares (EBSERH) (grant number 403940/2021-4), and Universidade Federal de Pernambuco (UFPE).
Conflict of Interest
The authors declare that there is no conflict of interest that could be perceived as prejudicial to the impartiality of the reported research.
Informed Consent
Written informed consent was obtained from all participants or their legal representatives.
Author Contributions
AB, RL, and NS conceived the study and drafted the manuscript. AB, RL, TL, AB, LS, JA, CS, AD, and BG were responsible for data acquisition and manuscript review. AB, RL, TL, AB, LS, JA, CS, CL, AD, BG, NB, LW, JS, and NS contributed to the literature search and critically reviewed the paper. NS supervised the project. All authors contributed to the article and approved the submitted version.
Data Availability
The authors declare that data supporting the findings of this study are available within the article.
| References | ▴Top |
- Cui X, Cui Y, Shi L, Luan J, Zhou X, Han J. A basic understanding of Turner syndrome: Incidence, complications, diagnosis, and treatment. Intractable Rare Dis Res. 2018;7(4):223-228.
doi pubmed - Kruszka P, Addissie YA, Tekendo-Ngongang C, Jones KL, Savage SK, Gupta N, Sirisena ND, et al. Turner syndrome in diverse populations. Am J Med Genet A. 2020;182(2):303-313.
doi pubmed - Bispo AV, Dos Santos LO, Buregio-Frota P, Galdino MB, Duarte AR, Leal GF, Araujo J, et al. Effect of chromosome constitution variations on the expression of Turner phenotype. Genet Mol Res. 2013;12(4):4243-4250.
doi pubmed - Gravholt CH, Andersen NH, Christin-Maitre S, Davis SM, Duijnhouwer A, Gawlik A, Maciel-Guerra AT, et al. Clinical practice guidelines for the care of girls and women with Turner syndrome. Eur J Endocrinol. 2024;190(6):G53-G151.
doi pubmed - Gravholt CH, Viuff MH, Brun S, Stochholm K, Andersen NH. Turner syndrome: mechanisms and management. Nat Rev Endocrinol. 2019;15(10):601-614.
doi pubmed - Gawlik AM, Berdej-Szczot E, Blat D, Klekotka R, Gawlik T, Blaszczyk E, Hankus M, et al. Immunological profile and predisposition to autoimmunity in girls with turner syndrome. Front Endocrinol (Lausanne). 2018;9:307.
doi pubmed - De Sanctis V, Khater D. Autoimmune diseases in Turner syndrome: an overview. Acta Biomed. 2019;90(3):341-344.
doi pubmed - Casto C, Pepe G, Li Pomi A, Corica D, Aversa T, Wasniewska M. Hashimoto's thyroiditis and Graves' disease in genetic syndromes in pediatric age. Genes (Basel). 2021;12(2):222.
doi pubmed - Santos LOD, Bispo AVS, Barros JV, Laranjeira RSM, Pinto RDN, Silva JA, Duarte AR, et al. CTLA-4 gene polymorphisms are associated with obesity in Turner Syndrome. Genet Mol Biol. 2018;41(4):727-734.
doi pubmed - Laranjeira RSM, Santos LO, Borborema MEA, Silva RFA, Bispo AVS, Duarte AR, Araujo J, et al. Upregulation of FOXP3 may act on immunological misbalance in Turner syndrome. Scand J Immunol. 2022;97:e13180.
doi - Mitsuiki N, Schwab C, Grimbacher B. What did we learn from CTLA-4 insufficiency on the human immune system? Immunol Rev. 2019;287(1):33-49.
doi pubmed - Garcia-Perez JE, Baxter RM, Kong DS, Tobin R, McCarter M, Routes JM, Verbsky J, et al. CTLA4 message reflects pathway disruption in monogenic disorders and under therapeutic blockade. Front Immunol. 2019;10:998.
doi pubmed - Verma N, Burns SO, Walker LSK, Sansom DM. Immune deficiency and autoimmunity in patients with CTLA-4 (CD152) mutations. Clin Exp Immunol. 2017;190(1):1-7.
doi pubmed - Khan U, Ghazanfar H. T lymphocytes and autoimmunity. Int Rev Cell Mol Biol. 2018;341:125-168.
doi pubmed - Bufalo NE, Dos Santos RB, Rocha AG, Teodoro L, Romaldini JH, Ward LS. Polymorphisms of the genes CTLA4, PTPN22, CD40, and PPARG and their roles in Graves' disease: susceptibility and clinical features. Endocrine. 2021;71(1):104-112.
doi pubmed - Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101-1108.
doi pubmed - Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611-622.
doi pubmed - https://www.snpstats.net/start.htm.
- https://www.r-project.org/.
- http://www.gpower.hhu.de/.
- Grossi A, Crino A, Luciano R, Lombardo A, Cappa M, Fierabracci A. Endocrine autoimmunity in Turner syndrome. Ital J Pediatr. 2013;39:79.
doi pubmed - Fang W, Zhang Z, Zhang J, Cai Z, Zeng H, Chen M, Huang J. Association of the CTLA4 gene CT60/rs3087243 single-nucleotide polymorphisms with Graves' disease. Biomed Rep. 2015;3(5):691-696.
doi pubmed - Tu Y, Fan G, Dai Y, Zeng T, Xiao F, Chen L, Kong W. Association between rs3087243 and rs231775 polymorphism within the cytotoxic T-lymphocyte antigen 4 gene and Graves' disease: a case/control study combined with meta-analyses. Oncotarget. 2017;8(66):110614-110624.
doi pubmed - Zahiri L, Malekmakan L, Masjedi F, Habibagahi Z, Habibagahi M. Association between IL-17A, FOXP3, and CTLA4 genes expression and severity of lupus nephritis. Iran J Kidney Dis. 2022;16(1):13-23.
doi pubmed - Royo JL. Hardy Weinberg equilibrium disturbances in case-control studies lead to non-conclusive results. Cell J. 2021;22(4):572-574.
doi pubmed - Catassi C, Verdu EF, Bai JC, Lionetti E. Coeliac disease. Lancet. 2022;399(10344):2413-2426.
doi pubmed
This article is distributed under the terms of the Creative Commons Attribution Non-Commercial 4.0 International License, which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Journal of Endocrinology and Metabolism is published by Elmer Press Inc.